Plotting
plotting.RmdManhattan
Here is a simple example of using the manhattan() plot
function.
library(genepi.utils)
gwas <- generate_random_gwas_data(100000)
highlight_snps <- gwas[["SNP"]][[which.min(gwas[["P"]])]]
annotate_snps <- gwas[["SNP"]][[which.min(gwas[["P"]])]]
p <- manhattan(gwas,
highlight_snps = highlight_snps,
highlight_win = 250,
annotate_snps = annotate_snps,
hit_table = TRUE,
title = "Manhattan Plot",
subtitle = "Example 1")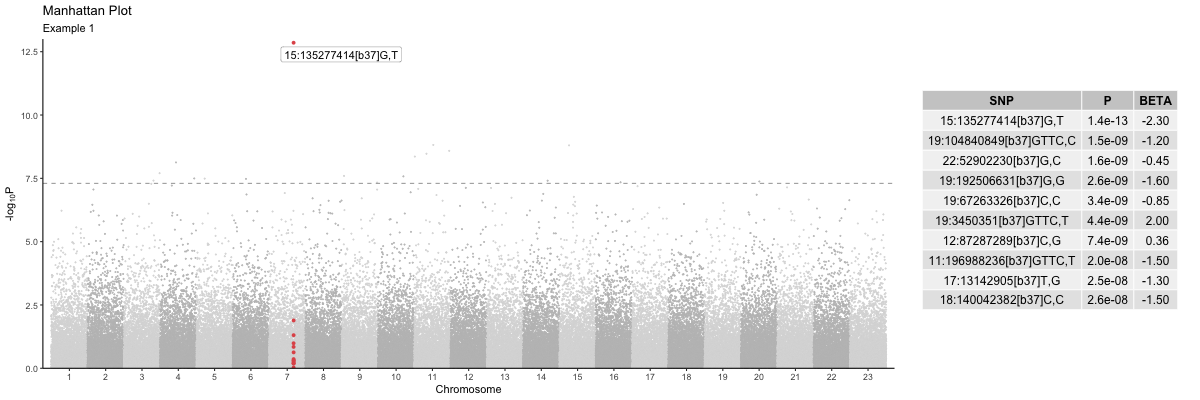
Miami
Here is a simple example of using the miami() plot
function. The use of named lists (i.e. ‘top’ and ‘bottom’) for the
parameters is not strictly necessary, but helps readability.
library(genepi.utils)
gwas_top <- generate_random_gwas_data(100000)
gwas_bottom <- generate_random_gwas_data(100000)
highlight_snps_top <- gwas[["SNP"]][[which.min(gwas[["P"]])]]
highlight_snps_botttom <- gwas[["SNP"]][[which.max(gwas[["P"]])]]
annotate_snps_top <- gwas[["SNP"]][[which.min(gwas[["P"]])]]
annotate_snps_botttom <- gwas[["SNP"]][[which.max(gwas[["P"]])]]
colours_top <- c("#67A3D9","#C8E7F5")
colours_bottom <- c("#F8B7CD","#F6D2E0")
p <- miami(gwases = list("top"=gwas_top, "bottom"=gwas_bottom),
highlight_snps = list("top"=highlight_snps_top, "bottom"=highlight_snps_botttom),
highlight_win = list("top"=250,"bottom"=250),
annotate_snps = list("top"=annotate_snps_top, "bottom"=annotate_snps_botttom),
colours = list("top"=colours_top, "bottom"=colours_bottom),
downsample = 0.0,
hit_table = TRUE,
title = "Miami Plot",
subtitle = list("top"="A", "bottom"="B"))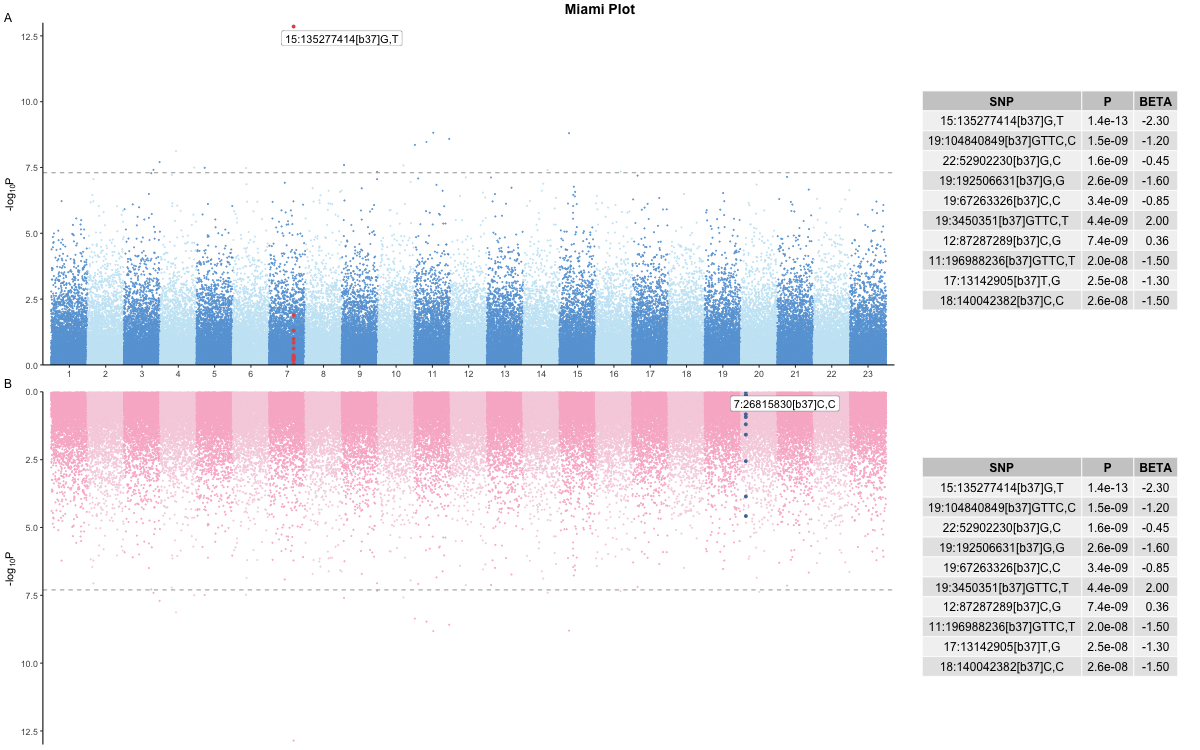
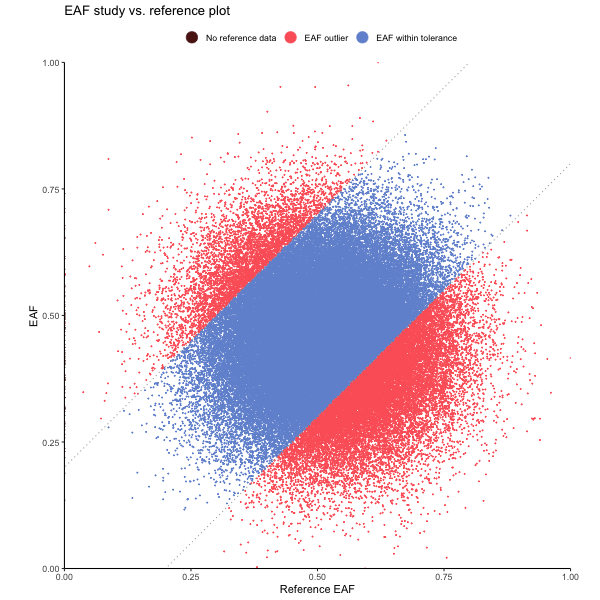
EAF plot
library(genepi.utils)
gwas <- generate_random_gwas_data(100000)
gwas[1:100, EUR_EAF := NA]
p <- eaf_plot(gwas,
eaf_col = "EAF",
ref_eaf_col = "EUR_EAF",
tolerance = 0.2,
colours = list(missing="#5B1A18", outlier="#FD6467", within="#7294D4"),
title = "EAF study vs. reference plot")